A Cambrian explosion of Scientific AI is unfolding, as once-separate components of scientific discovery become interoperable, API-accessible, and agent-operable. From molecular simulations to lab automation, from protein generation to literature analysis, the entire research stack is being digitized and wrapped in interfaces that LLMs and agents can interact with directly.
The implications are profound: algorithms autonomously generate hypotheses, design experiments, run simulations, and direct real-world assays. Closed-loop systems accelerate development cycles for therapeutics, materials, and beyond. As this ecosystem matures, an integrated map of capabilities and tools emerges, revealing a layered architecture of scientific AI that spans foundational models, orchestration, and human-facing platforms.
The Scientific AI Ecosystem Map aims to help founders, researchers and builders navigate this fast-moving frontier. We’ve organized the map into five key layers: (1) Knowledge & Data Sources, (2) Foundation & Domain Models, (3) Agentic Orchestration, (4) Experiment & Simulation Execution, and (5) Scientist-Facing Platforms & Assistants.
Below we’ll break down each layer and highlight key players driving innovation, ending with a look at how blockchain can unlock new levels of interoperability and decentralization - bridging isolated islands of the scientific AI landscape, encouraging wider and more diverse participation in the scientific process and fast-tracking discovery of frontier biotech.
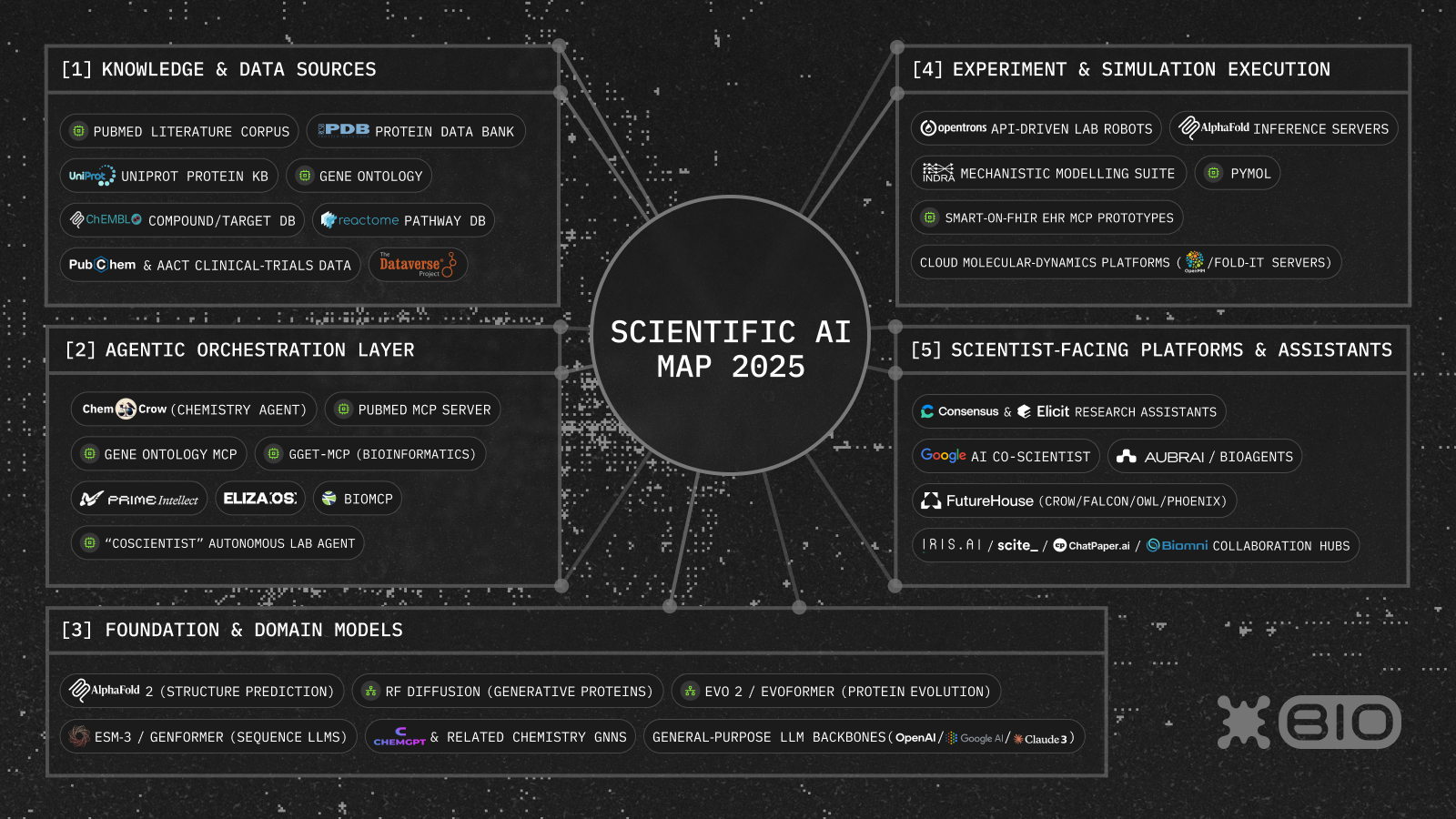
1. Knowledge & Data Sources
At the base are the vast knowledge and data repositories that AI systems feed on. Biology and chemistry have an explosion of publicly available data: over 25 million abstracts and 3 million full-text papers on PubMed, with ~4,000 new biomedical papers every day. Tens of thousands of protein structures live in the Protein Data Bank, millions of chemical compounds and reactions in databases like ChEMBL and PubChem, and curated ontologies (Gene Ontology, Reactome) organize gene and pathway knowledge.
Integrating and querying this sea of heterogeneous data is critical: early projects like FutureHouse have built crawlers to flag contradictions in the literature (they found ~2% of papers disagree with others, while automated knowledge graphs transform text into causal models. In short, these databases are the raw ingredients for AI-driven science.
Key examples:
- PubMed – the central biomedical literature database (25M+ abstracts)
- Dataverse Project – open-source research repository
- Protein Data Bank (PDB) – repository of 3D structures for proteins and nucleic acids.
- UniProt – a comprehensive protein sequence and functional annotation database.
- ChEMBL – curated bioactive drug-like molecules and targets.
- Gene Ontology – a standardized vocabulary of gene functions.
- Reactome – a curated database of biological pathways and reactions.
- PubChem – open repository of chemical structures and properties.
- AACT – the “Aggregate Analysis of ClinicalTrials.gov” relational database with every registered clinical study (protocol and results)
- bioRxiv – preprint server for biology
Together, these knowledge & data sources form the digital substrate of scientific AI. Language models and agents draw on them to answer questions, design experiments, and build virtual simulations.
2. Foundation & Domain Models
On top of the data layer sit the foundation models and domain-specific AI that can interpret it. These include both general-purpose LLMs and specialized models for biology/chemistry. For molecular structures, AlphaFold 2 (DeepMind) is a revolution – it predicts protein folds at near-experimental accuracy. The Baker lab’s RFdiffusion is a diffusion-based protein generator: a guided generative model that can design new protein backbones with high success (it outperformed prior design methods across many tasks).
There are also evolutionary protein language models (Meta’s ESM-3, Google’s Genformer) that understand sequence patterns, and “ChemGPT” or graph neural networks for chemistry that can predict reactions or material properties from raw inputs. These deep models are complemented by massive general LLMs – GPT-4, Claude 3, PaLM 2, etc. – which bring broad reasoning and language skills. The combination is potent: for example, in the Coscientist system at CMU, GPT-4 was one of several LLMs that collaborated to autonomously design and execute complex chemical syntheses.
Key examples:
- AlphaFold 2 (DeepMind) – AI for protein 3D structure prediction (CASP14 winner, near-experimental accuracy)
- RFdiffusion (Baker Lab) – a generative diffusion model that creates novel protein designs.
- Evoformer/Evo2 (AlphaFold’s internal evolution transformer) – algorithms that capture protein family evolution.
- ESM-3/Genformer (Meta) – large language models trained on protein sequences for structure & function prediction.
- ChemGPT & Chemistry GNNs – LLMs/graph networks tailored to chemical molecule representation and property prediction.
- General LLM backbones – GPT-4-class, Google’s PaLM 2, Anthropic’s Claude 3, etc., which provide the broad reasoning and coding skills underlying many agentic systems.
These foundation and domain models serve as the workhorses of the ecosystem. They transform raw sequences, structures, and molecular graphs into actionable insights – from folding prediction to retrosynthesis proposals – enabling the higher-level agents and tools described next.
3. Agentic Orchestration Layer
Above the models comes the orchestration – the AI agents and frameworks that coordinate tools, reason, and drive workflows. This layer glues together models, data sources, and execution platforms into cohesive “AI scientists.” For example, ChemCrow is a GPT-4–powered chemistry agent that chains together dozens of specialized tools (literature search, retrosynthesis, cost lookup, functional-group analysis, etc.) to autonomously tackle organic synthesis and drug design tasks. On the knowledge side, emerging MCP servers connect LLMs to biomedical databases. “BioMCP” is an open interface so agents can query genomics, drug, and pathway data via the Anthropic-led Model Context Protocol. Related servers like PubMed-MCP or Gene-Ontology-MCP let agents fetch specific literature annotations and ontology data on demand. Likewise, GGET-MCP is a bioinformatics MCP server (built on the gget library) that lets AI assistants perform gene and sequence analysis via natural language. In short, MCP servers are like API bridges so AI agents can call complex scientific tools securely and in a standardized way.
On the execution side, frameworks like Eliza OS provide a platform to run multiple AI agents and plugins. Eliza OS bills itself as a “Web3-friendly AI agent operating system”, allowing researchers to compose, deploy, and manage agent workflows on chain or locally. Teams like Prime Intellect are building decentralized compute fabrics so models and agents can be trained on distributed GPUs and co-owned. And research prototypes like Coscientist show how an autonomous agent can plan and run real chemistry experiments.
Key examples:
- ChemCrow & ChemMCP Toolkit – GPT-4 chemistry agent (Stanford) with ~18 expert tools for synthesis and analysis.
- BioMCP / PubMed-MCP / Gene-Ontology-MCP servers – interfaces exposing biomedical databases (genes, pathways, literature) via the Anthropic Model Context Protocol.
- elizaOS – a Web3 agent operating system.
- Prime Intellect – a decentralized protocol for pooling compute and training models globally.
- “Coscientist” AI lab agent – Carnegie Mellon’s GPT-4–driven system that autonomously designs and carries out complex chemistries in the lab.
- GGET-MCP – a bioinformatics MCP server (from the longevity-genie project) that exposes gene/sequence search, BLAST, AlphaFold, enrichment analysis, etc., via a standardized API
These agentic layers are the conductors: they orchestrate models, data, and robots. They break big problems into sub-steps, call the right tools, and handle error-checking and adaptation.
4. Experiment & Simulation Execution
The next layer executes experiments and simulations, closing the loop of scientific discovery. This includes lab automation, simulation engines, and other software platforms. On the wetlab side, open-source robotics kits like Opentrons let researchers script pipetting and lab processes via API, turning notebooks into lab robots. Cloud labs (e.g. Emerald Cloud Lab, Strateos) allow running bio and chem assays on demand from your laptop. On the dry side, software like INDRA is building mechanistic models: INDRA (Integrated Network and Dynamical Reasoning Assembler) reads mechanistic assertions from text/databases and composes causal networks and dynamical models.
Graphics engines like PyMOL and ready-to-run AlphaFold inference servers let scientists (and agents) visualize and refine structures. Healthcare data is also moving in: SMART-on-FHIR APIs and prototype MCP connectors are beginning to let AI agents query EHRs and clinical trial records in natural language. And molecular simulation platforms (OpenMM for molecular dynamics, Foldit’s crowd-lab servers, etc.) can compute 3D folding, docking, and dynamic hypotheses.
Key examples:
- Opentrons – open-source liquid-handling robots programmable via Python API.
- INDRA – a framework that assembles causal/dynamical models from literature and database statements.
- PyMOL / AlphaFold servers – tools for molecular visualization and on-demand structure prediction.
- SMART-on-FHIR EHR-MCP prototypes – emerging interfaces to query electronic health records.
- Cloud molecular simulation (e.g. OpenMM, Foldit servers) – platforms for protein folding and dynamics simulations.
This execution layer is the “body” of AI science: it physically or virtually runs the experiments that new hypotheses require. By automating lab protocols and simulations, it makes the feedback loop between AI planning and real (or simulated) experiments fast and scalable.
5. Scientist-Facing Platforms & Assistants
Finally, on top of everything are the interfaces and AI assistants that scientists use directly. These products often package many of the above tools into user-friendly apps or chatbots. For literature review, platforms like Consensus and Elicit let users ask biomedical questions and get summarized answers or relevant papers. Google’s AI Co-Scientist is a forthcoming assistant for literature search and analysis. Labs like FutureHouse (the developers behind Crow, Falcon, Owl, Phoenix) are building “multi-agent assistants” that can crawl data and write analyses. Blockchain-enabled agentic science like Aubrai and BioAgents are equipping scientific agents with Web3 features like decentralized storage and knowledge graphs. There are also specialized tools like Iris.ai for semantic paper search, Scite for evidence mapping of claims, ChatPaper.ai for AI-driven PDF chat, and unified agentic environments like Biomni.
Key Examples:
- Consensus & Elicit – AI-powered research assistants that synthesize answers from the literature.
- Google AI Co-Scientist – Google’s experimental research assistant for scientists.
- FutureHouse (Crow, Falcon, Owl, Phoenix) – a research lab building multi-agent assistants
- Aubrai / BioAgents – equipping scientific agents with onchain features.
- Iris.ai, Scite, ChatPaper.ai, Biomni – AI platforms and collaboration tools for scientific literature and data.
Conclusion
In the end, the true power of scientific AI comes from network effects. Despite its rapid progress, most scientific AI remains fragmented, unable to coordinate, transact, or build shared knowledge graphs decentrally. Bridging these islands is one of the next grand challenges to realize the full potential of scientific AI.
The more these tools interoperate, the faster hypotheses can be generated, tested, and shared. By introducing decentralized identity, data provenance, funding and incentives codified in smart contracts, we can enable transparent, tamper-resistant coordination and collaboration.
Imagine a global biology engine where humans and machines co-create knowledge in an open, secure and scalable manner. For a glimpse into Bio Protocol’s vision, check out this thread on BioAgents, and join the Bio community to stay up to date on our plans to bring scientific AI onchain.

.png)